Cytokine Signaling by the JAK STAT Pathway
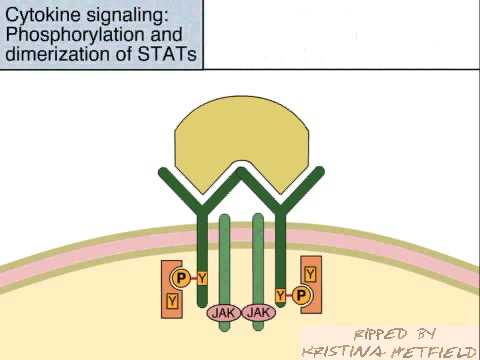
The JAK-STAT signaling pathway transmits information from chemical signals outside the cell, through the cell membrane, and into gene promoters on the DNA in the cell nucleus, which causes DNA transcription and activity in the cell. The JAK-STAT system is a major signaling alternative to the second messenger system. The JAK-STAT system consists of three main components: a receptor, JAK and STAT.
JAK is short for Janus Kinase, and STAT is short for Signal Transducer and Activator of Transcription.

The receptor is activated by a signal from interferon, interleukin, growth factors, or other chemical messengers. This activates the kinase function of JAK, which autophosphorylates itself (phosphate groups act as "on" and "off" switches on proteins). The STAT protein then binds to the phosphorylated receptor. STAT is phosphorylated and translocates into the cell nucleus, where it binds to DNA and promotes transcription of genes responsive to STAT.
In mammals, there are seven STAT genes, and each one binds to a different DNA sequence. STAT binds to a DNA sequence called a promoter, which controls the expression of other DNA sequences. This affects basic cell functions, like cell growth, differentiation and death.
The JAK-STAT pathway is evolutionarily conserved, from slime molds and worms to mammals (but not fungi or plants). Disrupted or dysregulated JAK-STAT functionality (which is usually by inherited or acquired genetic defects) can result in immune deficiency syndromes and cancers.
JAK is short for Janus Kinase, and STAT is short for Signal Transducer and Activator of Transcription.
The receptor is activated by a signal from interferon, interleukin, growth factors, or other chemical messengers. This activates the kinase function of JAK, which autophosphorylates itself (phosphate groups act as "on" and "off" switches on proteins). The STAT protein then binds to the phosphorylated receptor. STAT is phosphorylated and translocates into the cell nucleus, where it binds to DNA and promotes transcription of genes responsive to STAT.
In mammals, there are seven STAT genes, and each one binds to a different DNA sequence. STAT binds to a DNA sequence called a promoter, which controls the expression of other DNA sequences. This affects basic cell functions, like cell growth, differentiation and death.
The JAK-STAT pathway is evolutionarily conserved, from slime molds and worms to mammals (but not fungi or plants). Disrupted or dysregulated JAK-STAT functionality (which is usually by inherited or acquired genetic defects) can result in immune deficiency syndromes and cancers.
PI3K/AKT signaling pathway
Since its initial discovery as a proto-oncogene, the serine/threonine kinase Akt (also known as protein kinase B or PKB) has become a major focus of attention because of its critical regulatory role in diverse cellular processes, including cancer progression and insulin metabolism. The Akt cascade is activated by receptor tyrosine kinases, integrins, B and T cell receptors, cytokine receptors, G protein coupled receptors and other stimuli that induce the production of phosphatidylinositol 3,4,5 triphosphates (PtdIns(3,4,5)P3) by phosphoinositide 3-kinase (PI3K). These lipids serve as plasma membrane docking sites for proteins that harbor pleckstrin-homology (PH) domains, including Akt and its upstream activator PDK1. There are three highly related isoforms of Akt (Akt1, Akt2, and Akt3) and these represent the major signaling arm of PI3K.

For example, Akt is important for insulin signaling and glucose metabolism, with genetic studies in mice revealing a central role for Akt2 in these processes. Akt regulates cell growth through its effects on the mTOR and p70 S6 kinase pathways, as well as cell cycle and cell proliferation through its direct action on the CDK inhibitors p21 and p27, and its indirect effect on the levels of cyclin D1 and p53. Akt is a major mediator of cell survival through direct inhibition of pro-apoptotic signals such as Bad and the Forkhead family of transcription factors. T lymphocyte trafficking to lymphoid tissues is controlled by the expression of adhesion factors downstream of Akt. In addition, Akt has been shown to regulate proteins involved in neuronal function including GABA receptor, ataxin-1, and huntingtin proteins. Akt has been demonstrated to interact with Smad molecules to regulate TGFβ signaling. Finally, lamin A phosphorylation by Akt could play a role in the structural organization of nuclear proteins. These findings make Akt/PKB an important therapeutic target for the treatment of cancer, diabetes, laminopathies, stroke and neurodegenerative disease.
For example, Akt is important for insulin signaling and glucose metabolism, with genetic studies in mice revealing a central role for Akt2 in these processes. Akt regulates cell growth through its effects on the mTOR and p70 S6 kinase pathways, as well as cell cycle and cell proliferation through its direct action on the CDK inhibitors p21 and p27, and its indirect effect on the levels of cyclin D1 and p53. Akt is a major mediator of cell survival through direct inhibition of pro-apoptotic signals such as Bad and the Forkhead family of transcription factors. T lymphocyte trafficking to lymphoid tissues is controlled by the expression of adhesion factors downstream of Akt. In addition, Akt has been shown to regulate proteins involved in neuronal function including GABA receptor, ataxin-1, and huntingtin proteins. Akt has been demonstrated to interact with Smad molecules to regulate TGFβ signaling. Finally, lamin A phosphorylation by Akt could play a role in the structural organization of nuclear proteins. These findings make Akt/PKB an important therapeutic target for the treatment of cancer, diabetes, laminopathies, stroke and neurodegenerative disease.
Tarceva Mechanism Of Action
Erlotinib hydrochloride (originally coded as OSI-774) is a drug used to treat non-small cell lung cancer, pancreatic cancer and several other types of cancer.
Erlotinib specifically targets the epidermal growth factor receptor (EGFR) tyrosine kinase, which is highly expressed and occasionally mutated in various forms of cancer. It binds in a reversible fashion to the adenosine triphosphate (ATP) binding site of the receptor. For the signal to be transmitted, two members of the EGFR family need to come together to form a homodimer. These then use the molecule of ATP to autophosphorylate each other, which causes a conformational change in their intracellular structure, exposing a further binding site for binding proteins that cause a signal cascade to the nucleus. By inhibiting the ATP, autophosphorylation is not possible and the signal is stopped.
Erlotinib has shown a survival benefit in the treatment of lung cancer in phase III trials. It has been approved for the treatment of locally advanced or metastatic non-small cell lung cancer that has failed at least one prior chemotherapy regimen. In November 2005, the FDA approved the use of erlotinib in combination with gemcitabine for treatment of locally advanced, unresectable, or metastatic pancreatic cancer.
A test for the EGFR mutation in cancer patients has been developed by Genzyme. This may predict who will respond to erlotinib and other tyrosine kinase inhibitors. It is reported that responses among patients with lung cancer are seen most often in females who were never smokers, particularly Asian women and those with adenocarcinoma cell type.
Erlotinib has recently been shown to be a potent inhibitor of JAK2V617F activity. JAK2V617F is a mutant of tyrosine kinase JAK2, is found in most patients with polycythemia vera (PV) and a substantial proportion of patients with idiopathic myelofibrosis or essential thrombocythemia. The study suggests that erlotinib may be used for treatment of JAK2V617F-positive PV and other myeloproliferative disorders.
Microtubules
Microtubules are one of the components of the cytoskeleton. They have a diameter of 25 nm and length varying from 200 nanometers to 25 micrometers. Microtubules serve as structural components within cells and are involved in many cellular processes including mitosis, cytokinesis, and vesicular transport
Microtubules are polymers of α- and β-tubulin dimers. The tubulin dimers polymerize end to end in protofilaments. The protofilaments then bundle in hollow cylindrical filaments. Typically, the protofilaments arrange themselves in an imperfect helix with one turn of the helix containing 13 tubulin dimers each from a different protofilament. The image above illustrates a small section of microtubule, a few αβ dimers in length.
Another important feature of microtubule structure is polarity. Tubulin polymerizes end to end with the α subunit of one tubulin dimer contacting the β subunit of the next. Therefore, in a protofilament, one end will have the α subunit exposed while the other end will have the β subunit exposed. These ends are designated (−) and (+) respectively. The protofilaments bundle parallel to one another, so in a microtubule, there is one end, the (+) end, with only β subunits exposed while the other end, the (−) end, only has α subunits exposed.
Nucleation and growth
Polymerization of microtubules is nucleated in a microtubule organizing center. Contained within the MTOC is another type of tubulin, γ-tubulin, which is distinct from the α and β subunits which compose the microtubules themselves. The γ-tubulin combines with several other associated proteins to form a circular structure known as the "γ-tubulin ring complex." This complex acts as a scaffold for α/β tubulin dimers to begin polymerization; it acts as a cap of the (−) end while microtubule growth continues away from the MTOC in the (+) direction.
Ritonavir
Ritonavir, with trade name Norvir (Abbott Laboratories), is an antiretroviral drug from the protease inhibitor class used to treat HIV infection and AIDS.
Ritonavir is frequently prescribed with HAART, not for its antiviral action, but as it inhibits the same host enzyme that metabolizes other protease inhibitors. This inhibition leads to higher plasma concentrations of these latter drugs, allowing the clinician to lower their dose and frequency and improving their clinical efficacy.
Method of action
Ritonavir was originally developed as an inhibitor of HIV protease. It is one of the most complex inhibitors. It is now rarely used for its own antiviral activity, but remains widely used as a booster of other protease inhibitors. More specifically, ritonavir is used to inhibit a particular liver enzyme that normally metabolizes protease inhibitors, cytochrome P450-3A4 (CYP3A4).[3] The drug's molecular structure inhibits CYP3A4, so a low dose can be used to enhance other protease inhibitors. This discovery, which has drastically reduced the adverse effects and improved the efficacy of PI's and HAART, was first communicated in an article published in the AIDS Journal in 1997 by the University of Liverpool. This effect does come with a price: it also affects the efficacy of numerous other medications, making it difficult to know how to administer them concurrently. In addition it can cause a large number of side-effects on its own.
Drug interactions
Concomitant therapy of ritonavir with a variety of medications may result in serious and sometimes fatal drug interactions.[4] These interactions can occur with strong inhibitors, strong or moderate inducers or substrates of hepatic cytochrome P450 CYP3A4 isoform.
The list of clinically significant interactions of ritonavir includes but is not limited to following drugs:
- amiodarone - decreased metabolism, possible toxicity
- midazolam and triazolam - contraindicated
- carbamazepine - decreased metabolism, possible toxicity
- cisapride - decreased metabolism, possible prolongation of Q-T interval and life-threatening arrythmias
- disulfiram (with ritonavir oral preparation) - decreased metabolism of ritonavir
- eplerenone
- etravirine
- flecainide - decreased metabolism, possible toxicity
- MDMA
- meperidine - build-up of toxic concentrations of a metabolite possible
- nilotinib
- nisoldipine
- pimozide
- quinidine
- ranolazine
- salmeterol
- St John's wort
- statins - decreased metabolism, without dosage modification increased risk of rhabomyolisis
- thioridazine
- topotecan
- voriconazole - ritonavir increases metabolism of voriconazole
Mode of Action of Tipranavir in HIV
Tipranavir, or tipranavir disodium, is a nonpeptidic protease inhibitor (PI) manufactured by Boehringer-Ingelheim under the trade name Aptivus. It is administered with ritonavir in combination therapy to treat HIV infection and is given as two 250 mg capsules together with 200 mg of ritonavir twice daily.
Tipranavir has the ability to inhibit the replication of viruses that are resistant to other protease inhibitors and it recommended for patients who are resistant to other treatments. Resistance to tipranavir itself seems to require multiple mutations
Sarcomere Contraction Animation
A sarcomere is the basic unit of a muscle's cross-striated myofibril. Sarcomeres are multi-protein complexes composed of three different filament systems.
- The thick filament system is composed of myosin protein which is connected from the M-line to the Z-disc by Titin It also contains myosin-binding protein C which binds at one end to the thick filament and the other to Actin.
- The thin filaments are assembled by actin monomers bound to Nebulin. Which also involves tropomyosin; a dimer which coils itself around the F-actin core of the thin filament.
- Nebulin and Titin gives stability and structure to the sarcomere.
A muscle cell, from a biceps, may contain 100,000 sarcomeres. The myofibrils of smooth muscle cells are not arranged into sarcomeres.
Bands The sarcomeres are what give skeletal and cardiac muscles their striated appearance.
- A sarcomere is defined as the segment between two neighbouring Z-lines (or Z-discs, or Z bodies). In electron micrographs of cross striated muscle the Z-line (from the German "Zwischenscheibe", the band in between the I bands) appears as a series of dark lines.
- Surrounding the Z-line is the region of the I-band (for isotropic).
- Following the I-band is the A-band (for anisotropic). Named for their properties under a polarizing microscope.
- Within the A-band is a paler region called the H-band (from the German "Heller", bright). Named for their properties under a polarization microscope.
- Finally, inside the H-band is a thin M-line (from the German "Mittel", middle of the sarcomere).
- Actin filaments are the major component of the I-band and extend into the A-band.
- Myosin filaments extend throughout the A-band and are thought to overlap in the M-band.
- The giant protein titin (connectin) extends from the Z-line of the sarcomere, where it binds to the thin filament system, to the M-band, where it is thought to interact with the thick filaments. Titin (and its splice isoforms) is the biggest single highly elasticated protein found in nature. It provides binding sites for numerous proteins and is thought to play an important role as sarcomeric ruler and as blueprint for the assembly of the sarcomere.
- Several proteins important for the stability of the sarcomeric structure are found in the Z-line as well as in the M-band of the sarcomere.
- Actin filaments and Titin molecules are cross-linked in the Z-disc via the Z-line protein alpha-Actinin.
- The M-band proteins myomesin as well as M-protein crosslink the thick filament system (myosins) and the M-band part of titin (the elastic filaments).
- The interaction between actin and myosin filaments in the A-band of the sarcomere is responsible for the muscle contraction (sliding filament model).
Upon muscle contraction, the A-bands do not change their length (1.85 micrometer in mammalian skeletal muscle) whereas the I-bands and the H-bands shorten.
The A-band, I-band and Z-line are never visible even at the light-microscope level. The protein actin myosin covers the myosin binding sites of the actin molecules in the muscle cell. To allow the muscle cell to contract, tropomyosin must be moved to uncover the binding sites on the actin. Calcium ions bind with troponin molecules (which are dispersed throughout the tropomyosin protein) and alter the structure of the tropomyosin, forcing it to reveal the cross bridge binding site on the actin. The concentration of calcium within muscle cells is controlled by the sarcoplasmic reticulum, a unique form of endoplasmic reticulum. Muscle contraction ends when calcium ions are pumped back out of the sarcomere.
Skeletal muscle never contracts even when an impulse is received from a motor neuron . During stimulation of the muscle cell, the motor neuron releases the neurotransmitter acetylcholine which travels across the neuromuscular junction (the synapse between the terminal bouton of the neuron and the muscle cell). The action potential then travels along T (transverse) tubules until it reaches the sarcoplasmic reticulum; the action potential from the motor neuron changes the permeability of the sarcoplasmic reticulum, allowing the flow of calcium ions into the sarcomere. The outflow of calcium allows the myosin heads access to the actin cross bridge binding sites, permitting muscle contraction.
Rest At rest, the myosin head is bound to an ATP molecule in a low-energy configuration and is unable to access the cross bridge binding sites on the actin. However, the myosin head can hydrolyze ATP into ADP and an inorganic phosphate ion. A portion of the energy released in this reaction changes the shape of the myosin head and promotes it to a high-energy configuration. Through the process of binding to the actin, the myosin head releases ADP and inorganic phosphate ion, changing its configuration back to one of low energy. The myosin remains attached to actin in a state known as Rigor, until a new ATP binds the myosin head. This binding of ATP to myosin releases the actin by cross-bridge dissociation. The ATP associated myosin is ready for another cycle, beginning with hydrolysis of the ATP.
Storage Most muscle cells only store enough ATP for a small number of muscle contractions. While muscle cells also store glycogen, most of the energy required for contraction is derived from phosphagens. One such phosphagen is creatine phosphate, which is used to provide ADP with a phosphate group for ATP synthesis in vertebrates.
RESTRICTION ENZYME

A restriction enzyme (or restriction endonuclease) is an enzyme that cuts double-stranded or single stranded DNA at specific recognition nucleotide sequences known as restriction sites. Such enzymes, found in bacteria and archaea, are thought to have evolved to provide a defense mechanism against invading viruses.Inside a bacterial host, the restriction enzymes selectively cut up foreign DNA in a process called restriction; host DNA is methylated by a modification enzyme (a methylase) to protect it from the restriction enzyme’s activity. Collectively, these two processes form the restriction modification system. cut the DNA, a restriction enzyme makes two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix.
Norovirus Animation

Norovirus, an RNA virus of the Caliciviridae taxonomic family, causes approximately 90% of epidemic non-bacterial outbreaks of gastroenteritis around the world, and is responsible for 50% of all foodborne outbreaks of gastroenteritis in the US. Norovirus affects people of all ages. The viruses are transmitted by faecally contaminated food or water and by person-to-person contact.
After infection, immunity to norovirus is not complete nor long-lasting. There is an inherited predisposition to infection and people whose blood type can be detected in their saliva are more often infected.
Outbreaks of norovirus disease often occur in closed or semi-closed communities, such as long-term care facilities, hospitals, prisons and cruise ships where once the virus has been introduced, the infection spreads very rapidly by either person-to-person transmission or through contaminated food.Many norovirus outbreaks have been traced to food that was handled by one infected person.
Norovirus is rapidly killed by chlorine-based disinfectants, but because the virus particle does not have a lipid envelope, it is less susceptible to alcohols and detergents.
There are different genogroups of norovirus and the majority of noroviruses that infect humans are classified into genogroup G1 and G2.
History
Originally, norovirus was named after Norwalk, Ohio, where an outbreak of acute gastroenteritis occurred among children at an elementary school in November 1968. In 1972, immune electron microscopy on stored stool samples identified a virus, which was given the name Norwalk virus. Numerous outbreaks with similar symptoms have been reported since. The cloning and sequencing of the Norwalk virus genome showed that these viruses have a genomic organization consistent with viruses belonging to the family Caliciviridae. The name norovirus (Norovirus for the genus) was approved by the International Committee on Taxonomy of Viruses in 2002.
Common names of the illness caused by noroviruses are stomach flu, winter vomiting disease, viral gastroenteritis and acute non-bacterial gastroenteritis.
Some previously used names which can be used for PubMed and other internet searches are Norwalk virus, Norwalk-like virus, SRSVs (Small Round Structured Viruses), Sapporo virus and Snow Mountain.
Signs and symptoms
The disease is usually self-limiting, and characterised by nausea, vomiting, diarrhea, and abdominal pain. General lethargy, weakness, muscle aches, headache, and low-grade fever may occur. Symptoms may persist for several days and may become life-threatening in the young, the elderly, and the immune-compromised if dehydration is ignored or not treated.
Diagnosis
Specific diagnosis of norovirus is routinely made by polymerase chain reaction (PCR) assays or real-time PCR assays, which give results within a few hours. These assays are very sensitive and can detect concentrations as low as 10 virus particles.
Tests such as EIA that use antibodies against a mixture of norovirus strains are available commercially but lack specificity and sensitivity.
Prevention and infection control
Hand washing remains an effective method to reduce the spread of norovirus pathogens. Norovirus can be aerosolized when those stricken with the illness vomit. Surface sanitizing is recommended in areas where the Norovirus may be present on surfaces.
In health care environments, the prevention of nosocomial infections involves routine and terminal cleaning. Nonflammable alcohol vapor in CO2 systems are used in health care environments where medical electronics would be adversely affected by aerosolized chlorine or other caustic compounds.
Associated foods
Noroviruses are transmitted directly via person to person or indirectly via contaminated water and foods. A CDC study of eleven outbreaks in New York State lists the suspected mode of transmission as person-to-person in seven outbreaks, foodborne in two, waterborne in one, and one unknown. The source of waterborne outbreaks may include water from municipal supplies, wells, recreational lakes, swimming pools and ice machines.
Shellfish and salad ingredients are the foods most often implicated in Norwalk outbreaks. Ingestion of raw or insufficiently steamed clams and oysters poses a high risk for infection with the Norwalk virus. Foods other than shellfish are contaminated by ill food handlers.
Microbiology
Classification
Noroviruses can genetically be classified into 5 different genogroups (GI, GII, GIII, GIV, and GV) which can be further divided into different genetic clusters or genotypes. For example genogroup II, the most prevalent human genogroup, presently contains 19 genotypes. Genogroups I, II and IV infect humans, whereas genogroup III infects bovine species and genogroup V has recently been isolated in mice.
Noroviruses from Genogroup II, genotype 4 (abbreviated as GII.4) account for the majority of adult outbreaks of gastroenteritis and often sweep across the globe. Recent examples include US95/96-US strain, associated with global outbreaks in the mid- to late-90s, Farmington Hills virus associated with outbreaks in Europe and the United States in 2002 and in 2004 Hunter virus was associated with outbreaks in Europe, Japan and Australasia. In 2006 there was another large increase in NoV infection around the globe. In December, 2007 there was an outbreak at a country club in northern California where around 80-100 people were infected. Two new GII.4 variants caused around 80% of those Norovirus associated outbreaks and they have been termed 2006a and 2006b.Recent reports have shown a link between blood group and susceptibility to infection by norovirus.
Virus structure
Noroviruses contain a positive-sense RNA genome of approximately 7.5 kbp, encoding a major structural protein (VP1) of about 58~60 kDa and a minor capsid protein (VP2).The virus particles demonstrate an amorphous surface structure when visualized using electron microscopy and are between 27-38 nm in size.
Krebs Cycle
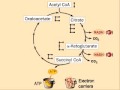
The citric acid cycle, also known as the tricarboxylic acid cycle (TCA cycle) or the Krebs cycle, (or rarely, the Szent-Györgyi-Krebs cycle) is a series of enzyme-catalysed chemical reactions of central importance in all living cells that use oxygen as part of cellular respiration. In eukaryotes, the citric acid cycle occurs in the matrix of the mitochondrion. The components and reactions of the citric acid cycle were established by seminal work from both Albert Szent-Györgyi and Hans Krebs.
In aerobic organisms, the citric acid cycle is part of a metabolic pathway involved in the chemical conversion of carbohydrates, fats and proteins into carbon dioxide and water to generate a form of usable energy. Other relevant reactions in the pathway include those in glycolysis and pyruvate oxidation before the citric acid cycle, and oxidative phosphorylation after it. In addition, it provides precursors for many compounds including some amino acids and is therefore functional even in cells performing fermentation.
A simplified view of the process The citric acid cycle begins with acetyl-CoA transferring its two-carbon acetyl group to the four-carbon acceptor compound (oxaloacetate) to form a six-carbon compound (citrate). The citrate then goes through a series of chemical transformations, losing first one, then a second carboxyl group as CO2. The carbons lost as CO2 originate from what was oxaloacetate, not directly from acetyl-CoA. The carbons donated by acetyl-CoA become part of the oxaloacetate carbon backbone after the first turn of the citric acid cycle. Loss of the acetyl-CoA-donated carbons as CO2 requires several turns of the citric acid cycle. However, because of the role of the citric acid cycle in anabolism, they may not be lost since many TCA cycle intermediates are also used as precursors for the biosynthesis of other molecules.
Most of the energy made available by the oxidative steps of the cycle is transferred as energy-rich electrons to NAD+, forming NADH. For each acetyl group that enters the citric acid cycle, three molecules of NADH are produced. Electrons are also transferred to the electron acceptor Q, forming QH2. At the end of each cycle, the four-carbon oxaloacetate has been regenerated, and the cycle continues.Products
Products of the first turn of the cycle are: one GTP (or ATP), three NADH, one QH2, two CO2.
Because two acetyl-CoA molecules are produced from each glucose molecule, two cycles are required per glucose molecule. Therefore, at the end of all cycles, the products are: two GTP, six NADH, two QH2, and four CO2
| Description | Reactants | Products |
| The sum of all reactions in the citric acid cycle is: | Acetyl-CoA + 3 NAD+ + Q + GDP + Pi + 2 H2O | → CoA-SH + 3 NADH + 3 H+ + QH2 + GTP + 2 CO2 |
| Combining the reactions occurring during the pyruvate oxidation with those occurring during the citric acid cycle, the following overall pyruvate oxidation reaction is obtained: | Pyruvic acid + 4 NAD+ + Q + GDP + Pi + 2 H2O | → 4 NADH + 4 H+ + QH2 + GTP + 3 CO2 |
| Combining the above reaction with the ones occurring in the course of glycolysis, the following overall glucose oxidation reaction (excluding reactions in the respiratory chain) is obtained: | Glucose + 10 NAD+ + 2 Q + 2 ADP + 2 GDP + 4 Pi + 2 H2O | → 10 NADH + 10 H+ + 2 QH2 + 2 ATP + 2 GTP + 6 CO2 |
The regulation of the TCA cycle is largely determined by substrate availability and product inhibition. NADH, a product of all dehydrogenases in the TCA cycle with the exception of succinate dehydrogenase, inhibits pyruvate dehydrogenase, isocitrate dehydrogenase and α-ketoglutarate dehydrogenase, and also citrate synthase. Acetyl-CoA inhibits pyruvate dehydrogenase, while succinyl-CoA inhibits succinyl-CoA synthase and citrate synthase. When tested in vitro with TCA enzymes, ATP inhibits citrate synthase and α-ketoglutarate dehydrogenase; however, ATP levels do not change more than 10% in vivo between rest and vigorous exercise. There is no known allosteric mechanism that can account for large changes in reaction rate from an allosteric effector whose concentration changes less than 10%
Calcium is used as a regulator. It activates pyruvate dehydrogenase, isocitrate dehydrogenase and α-ketoglutarate dehydrogenase. This increases the reaction rate of many of the steps in the cycle, and therefore increases flux throughout the pathway.
Citrate is used for feedback inhibition, as it inhibits phosphofructokinase, an enzyme involved in glycolysis that catalyses formation of fructose 1,6-bisphosphate, a precursor of pyruvate. This prevents a constant high rate of flux when there is an accumulation of citrate and a decrease in substrate for the enzyme.
Recent work has demonstrated an important link between intermediates of the citric acid cycle and the regulation of hypoxia inducible factors (HIF). HIF plays a role in the regulation of oxygen haemostasis, and is a transcription factor which targets angiogenesis, vascular remodelling, glucose ulitisation, iron transport and apoptosis. HIF is synthesized consititutively and hydroxylation of at least one of two critical proline residues mediates their interation with the von Hippel Lindau E3 ubiquitin ligase complex which targets them for rapid degradation. This reaction is calalysed by prolyl 4-hydroxylases. Fumarate and succinate have been identified as potent inhibitors of prolyl hydroxylases thus leading to the stabilisation of HIF.
Major metabolic pathways converging on the TCA cycle Several catabolic pathways converge on the TCA cycle. Reactions that form intermediates of the TCA cycle in order to replenish them (especially during the scarcity of the intermediates) are called anaplerotic reactions.
The citric acid cycle is the third step in carbohydrate catabolism (the breakdown of sugars). Glycolysis breaks glucose (a six-carbon-molecule) down into pyruvate (a three-carbon molecule). In eukaryotes, pyruvate moves into the mitochondria. It is converted into acetyl-CoA by decarboxylation and enters the citric acid cycle.
In protein catabolism, proteins are broken down by protease enzymes into their constituent amino acids. The carbon backbone of these amino acids can become a source of energy by being converted to Acetyl-CoA and entering into the citric acid cycle.
In fat catabolism, triglycerides are hydrolyzed to break them into fatty acids and glycerol. In the liver the glycerol can be converted into glucose via dihydroxyacetone phosphate and glyceraldehyde-3-phosphate by way of gluconeogenesis. In many tissues, especially heart tissue, fatty acids are broken down through a process known as beta oxidation which results in acetyl-CoA which can be used in the citric acid cycle. Sometimes beta oxidation can yield propionyl CoA which can result in further glucose production by gluconeogenesis in the liver.
The citric acid cycle is always followed by oxidative phosphorylation. This process extracts the energy (as electrons) from NADH and QH2, oxidizing them to NAD+ and Q, respectively, so that the cycle can continue. Whereas the citric acid cycle does not use oxygen, oxidative phosphorylation does.
The total energy gained from the complete breakdown of one molecule of glucose by glycolysis, the citric acid cycle and oxidative phosphorylation equals about 30 ATP molecules, in eukaryotes. The citric acid cycle is called an amphibolic pathway because it participates in both catabolism and anabolism.
Urinary Tract Infections (UTI)
A urinary tract infection (UTI) is a bacterial infection that affects any part of the urinary tract. Although urine contains a variety of fluids, salts, and waste products, it usually does not have bacteria in it. When bacteria get into the bladder or kidney and multiply in the urine, they cause a UTI. The most common type of UTI is a bladder infection which is also often called cystitis. Another kind of UTI is a kidney infection, known as pyelonephritis, and is much more serious. Although they cause discomfort, urinary tract infections can usually be quickly and easily treated when the patient sees a doctor promptly.
Symptoms & signs For bladder infections
- Frequent urination along with the feeling of having to urinate even though there may be very little urine to pass.
- Nocturia: Need to urinate during the night.
- Urethritis: Discomfort or pain at the urethral meatus or a burning sensation throughout the urethra with urination (dysuria).
- Pain in the midline suprapubic region.
- Pyuria: Pus in the urine or discharge from the urethra.
- Hematuria: Blood in urine.
- Pyrexia: Mild fever
- Cloudy and foul-smelling urine
- Increased confusion and associated falls are common presentations to Emergency Departments for elderly patients with UTI.
- Some urinary tract infections are asymptomatic.
- Protein found the urine.
- The above symptoms.
- Emesis: Vomiting is common.
- Back, side (flank) or groin pain.
- Abdominal pain or pressure.
- Shaking chills and high spiking fever.
- Night sweats.
- Extreme fatigue.
Since bacteria can enter the urinary tract through the urethra (an ascending infection), poor toilet habits can predispose to infection, but other factors (pregnancy in women, prostate enlargement in men) are also important and in many cases the initiating event is unclear.
While ascending infections are generally the rule for lower urinary tract infections and cystitis, the same may not necessarily be true for upper urinary tract infections like pyelonephritis which may be hematogenous in origin.
Allergies can be a hidden factor in urinary tract infections. For example, allergies to foods can irritate the bladder wall and increase susceptibility to urinary tract infections. Keep track of your diet and have allergy testing done to help eliminate foods that may be a problem. Urinary tract infections after sexual intercourse can be also be due to an allergy to latex condoms, spermicides, or oral contraceptives. In this case review alternative methods of birth control with your doctor.
The use of urinary catheters in both women and men who are elderly, people experiencing nervous system disorders and people who are convalescing or unconscious for long periods of time may result in an increased risk of urinary tract infection for a variety of reasons. Scrupulous aseptic technique may decrease this risk.
The bladder wall is coated with various mannosylated proteins, such as Tamm-Horsfall proteins (THP), which interfere with the binding of bacteria to the uroepithelium. As binding is an important factor in establishing pathogenicity for these organisms, its disruption results in reduced capacity for invasion of the tissues. Moreover, the unbound bacteria are more easily removed when voiding. The use of urinary catheters (or other physical trauma) may physically disturb this protective lining, thereby allowing bacteria to invade the exposed epithelium.
Elderly individuals, both men and women, are more likely to harbor bacteria in their genitourinary system at any time. These bacteria may be associated with symptoms and thus require treatment with an antibiotic. The presence of bacteria in the urinary tract of older adults, without symptoms or associated consequences, is also a well recognized phenomenon which may not require antibiotics. This is usually referred to as asymptomatic bacteriuria. The overuse of antibiotics in the context of bacteriuria among the elderly is a concerning and controversial issue.
Women are more prone to UTIs than males because in females, the urethra is much shorter and closer to the anus than in males, and they lack the bacteriostatic properties of prostatic secretions. Among the elderly, UTI frequency is in roughly equal proportions in women and men.
A common cause of UTI is an increase in sexual activity, such as vigorous sexual intercourse with a new partner. The term "honeymoon cystitis" has been applied to this phenomenon.
A neuroinformatics perspective on cerebral cortical structure and function

David Van Essen, Washington University, St. Louis, USA. Keynote lecture at Neuroinformatics 2008 in Stockholm, Sweden.
Ligand binding and action of microswitches

Ligand binding and action of microswitches in G protein coupled receptors. Slawomir Filipek, International Institute of Molecular and Cell Biology, Warsaw, Poland. Workshop lecture at Neuroinformatics 2010 in Kobe, Japan
Paramecium Dividing
Paramecia are unicellular organisms usually less than 0.25mm in length and covered with minute hair-like projections called cilia. They are characterized by their cilia which are used in locomotion and during feeding. Paramecia feed on bacteria.
Classification:
Kingdom – Protista
Phylum – Ciliophora
Order – Hymenostomatida
Family -- Paramecidae
Genus – Paramecium
Species – caudatum
Natural Habitat:
Fresh water
Why fresh water?
They take in water by osmosis from the hypotonic environment, bladder-like contractile vacuoles accumulate the excess water from radial canals and periodically expel it through the plasma membrane by contractions of the surrounding cytoplasm.
Interesting characteristics:
Paramecia have 2 nuclei, 1 macronucleus and 1 micronucleus. Some have up to 80 micronuclei!
The organism cannot survive without macronucleus and cannot reproduce without micronucleus.
The organism cannot survive without macronucleus and cannot reproduce without micronucleus.
Reproduction is either by asexual binary fission or occasionally by conjugation (sexual). During binary fission a fully grown organism divides into two daughter cells. Conjugation consists of the temporary union of 2 organisms and the exchange of micronuclear elements. Without the rejuvenating effects of conjugation a paramecium ages and dies. Only opposite mating types, or genetically compatible organisms, can unite in conjugation
Paramecium caudatum in conjugation

Paramecium caudatum are unicellular organisms belonging to the genus of protozoa of the phylum Ciliophora. They are less than 0.25mm in length and covered with minute hair-like projections called cilia. The cilia are used in locomotion and during feeding. They are often called slipper animalcules because of their slipper-like shape.
Paramecium have 2 nuclei (a large macronucleus and a single compact micronucleus). They cannot survive without macro-nucleus and cannot reproduce without micro-nucleus. Reproduction is either by asexual binary fission or occasionally by conjugation (sexual) and rarely by endomixis, a process involving total nuclear reorganization of individual organisms. During binary fission a fully grown organism divides into two daughter cells. Conjugation consists of the temporary union of 2 organisms and the exchange of micro-nuclear elements. Without the rejuvenating effects of conjugation a paramecium ages and dies. Only opposite mating types, or genetically compatible organisms, can unite in conjugation.
Ribosome

Ribosomes (from ribonucleic acid and "greek: soma (meaning body)") are complexes of RNA and protein that are found in all cells. Prokaryotic ribosomes from archaea and bacteria are smaller than most of the ribosomes from eukaryotes such as plants and animals. However, the ribosomes in the mitochondrion of eukaryotic cells resemble those in bacteria, reflecting the evolutionary origin of this organelle.
The function of ribosomes is the assembly of proteins, in a process called translation. Ribosomes do this by catalysing the assembly of individual amino acids into polypeptide chains; this involves binding a messenger RNA and then using this as a template to join together the correct sequence of amino acids. This reaction uses adapters called transfer RNA molecules, which read the sequence of the messenger RNA and are attached to the amino acids.
Ribosomes are the workhorses of protein biosynthesis, the process of translating RNA into protein. The mRNA comprises a series of codons that dictate to the ribosome the sequence of the amino acids needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon of the mRNA, pairing it with the appropriate amino acid. This is done using molecules of transfer RNA (tRNA) containing a complementary anticodon on one end and the appropriate amino acid on the other.
Protein synthesis begins at a start codon near the 5' end of the mRNA. The small ribosomal subunit, typically bound to a tRNA containing the amino acid methionine, binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. The large ribosomal subunit contains three tRNA binding sites, designated A, P, and E. The A site binds an aminoacyl-tRNA (a tRNA bound to an amino acid); the P site binds a peptidyl-tRNA (a tRNA bound to the peptide being synthesized); and the E site binds a free tRNA before it exits the ribosome.
In Figure 3, both ribosomal subunits (small and large) assemble at the start codon (towards the 5' end of the mRNA). The ribosome uses tRNA which matches the current codon (triplet) on the mRNA to append an amino acid to the polypeptide chain. This is done for each triplet on the mRNA, while the ribosome moves towards the 3' end of the mRNA. Usually in bacterial cells, several ribosomes are working parallel on a single mRNA, forming what we call a polyribosome or polysome.
Ribosomes are the workhorses of protein biosynthesis, the process of translating RNA into protein. The mRNA comprises a series of codons that dictate to the ribosome the sequence of the amino acids needed to make the protein. Using the mRNA as a template, the ribosome traverses each codon of the mRNA, pairing it with the appropriate amino acid. This is done using molecules of transfer RNA (tRNA) containing a complementary anticodon on one end and the appropriate amino acid on the other.
Protein synthesis begins at a start codon near the 5' end of the mRNA. The small ribosomal subunit, typically bound to a tRNA containing the amino acid methionine, binds to an AUG codon on the mRNA and recruits the large ribosomal subunit. The large ribosomal subunit contains three tRNA binding sites, designated A, P, and E. The A site binds an aminoacyl-tRNA (a tRNA bound to an amino acid); the P site binds a peptidyl-tRNA (a tRNA bound to the peptide being synthesized); and the E site binds a free tRNA before it exits the ribosome.
In Figure 3, both ribosomal subunits (small and large) assemble at the start codon (towards the 5' end of the mRNA). The ribosome uses tRNA which matches the current codon (triplet) on the mRNA to append an amino acid to the polypeptide chain. This is done for each triplet on the mRNA, while the ribosome moves towards the 3' end of the mRNA. Usually in bacterial cells, several ribosomes are working parallel on a single mRNA, forming what we call a polyribosome or polysome.
Bacterial conjugation

Bacterial conjugation is the transfer of genetic material between bacteria through direct cell-to-cell contact. Discovered in 1946 by Joshua Lederberg and Edward Tatum,conjugation is a mechanism of horizontal gene transfer—as are transformation and transduction—although these mechanisms do not involve cell-to-cell contact.
Bacterial conjugation is often incorrectly regarded as the bacterial equivalent of sexual reproduction or mating. It is not actually sexual, as it does not involve the fusing of gametes and the creation of a zygote, nor is there equal exchange of genetic material. It is merely the transfer of genetic information from a donor cell to a recipient. In order to perform conjugation, one of the bacteria, the donor, must play host to a conjugative or mobilizable genetic element, most often a conjugative or mobilizable plasmid or transposon. Most conjugative plasmids have systems ensuring that the recipient cell does not already contain a similar element.
The genetic information transferred is often beneficial to the recipient cell. Benefits may include antibiotic resistance, other xenobiotic tolerance, or the ability to utilize a new metabolite. Such beneficial plasmids may be considered bacterial endosymbionts. Some conjugative elements may also be viewed as genetic parasites on the bacterium, and conjugation as a mechanism was evolved by the mobile element to spread itself into new hosts.
Mechanism
The prototype for conjugative plasmids is the F-plasmid, also called the F-factor. The F-plasmid is an episome (a plasmid that can integrate itself into the bacterial chromosome by genetic recombination) of about 100 kb length. It carries its own origin of replication, the oriV, as well as an origin of transfer, or oriT. There can only be one copy of the F-plasmid in a given bacterium, either free or integrated (two immediately before cell division). The host bacterium is called F-positive or F-plus (denoted F+). Strains that lack F plasmids are called F-negative or F-minus (F-).
Among other genetic information, the F-plasmid carries a tra and a trb locus, which together are about 33 kb long and consist of about 40 genes. The tra locus includes the pilin gene and regulatory genes, which together form pili on the cell surface, polymeric proteins that can attach themselves to the surface of F- bacteria and initiate the conjugation. Though there is some debate on the exact mechanism, the pili themselves do not seem to be the structures through which the actual exchange of DNA takes place; rather, some proteins coded in the tra or trb loci seem to open a channel between the bacteria.
When conjugation is initiated, via a mating signal, a relaxase enzyme creates a nick in one plasmid DNA strand at the origin of transfer, or oriT. The relaxase may work alone or in a complex of over a dozen proteins, known collectively as a relaxosome. In the F-plasmid system, the relaxase enzyme is called TraI and the relaxosome consists of TraI, TraY, TraM, and the integrated host factor, IHF. The transferred, or T-strand, is unwound from the duplex plasmid and transferred into the recipient bacterium in a 5'-terminus to 3'-terminus direction. The remaining strand is replicated, either independent of conjugative action (vegetative replication, beginning at the oriV) or in concert with conjugation (conjugative replication similar to the rolling circle replication of lambda phage). Conjugative replication may necessitate a second nick before successful transfer can occur. A recent report claims to have inhibited conjugation with chemicals that mimic an intermediate step of this second nicking event.
If the F-plasmid becomes integrated into the host genome, donor chromosomal DNA may be transferred along with plasmid DNA. The certain amount of chromosomal DNA that is transferred depends on how long the bacteria remain in contact; for common laboratory strains of E. coli the transfer of the entire bacterial chromosome takes about 100 minutes. The transferred DNA can be integrated into the recipient genome via recombination.
A culture of cells containing non-integrated F plasmids usually contains a few that have accidentally become integrated, and these are responsible for those low-frequency chromosomal gene transfers which do occur in such cultures. Some strains of bacteria with an integrated F-plasmid can be isolated and grown in pure culture. Because such strains transfer chromosomal genes very efficiently, they are called Hfr (high frequency of recombination). The E. coli genome was originally mapped by interrupted mating experiments, in which various Hfr cells in the process of conjugation were sheared from recipients after less than 100 minutes (initially using a Waring blender) and investigating which genes were transferred.
DNA STRUCTURE

In 1951, the then 23-year old biologist James Watson traveled from the United States to work with Francis Crick, an English physicist at the University of Cambridge. Crick was already using the process of X-ray crystallography to study the structure of protein molecules. Together, Watson and Crick used X-ray crystallography data, produced by Rosalind Franklin and Maurice Wilkins at King's College in London, to decipher DNA's structure.
- This is what they already knew from the work of many scientists, about the DNA molecule:
- DNA is made up of subunits which scientists called nucleotides.
- Each nucleotide is made up of a sugar, a phosphate and a base.
- There are 4 different bases in a DNA molecule:
adenine (a purine)
cytosine (a pyrimidine)
guanine (a purine)
Thymine (a pyrimidine)
- The number of purine bases equals the number of pyrimidine bases
- The number of adenine bases equals the number of thymine bases
- The number of guanine bases equals the number of cytosine bases
- The basic structure of the DNA molecule is helical, with the bases being stacked on top of each other
Working with nucleotide models made of wire, Watson and Crick attempted to put together the puzzle of DNA structure in such a way that their model would account for the variety of facts that they knew described the molecule. Once satisfied with their model, they published their hypothesis, entitled "Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid" in the British journal Nature (April 25, 1953. volume 171:737-738.) It is interesting to note that this paper has been cited over 800 times since its first appearance!
Here are their words:
"...This (DNA) structure has two helical chains each coiled round the same axis...Both chains follow right handed helices...the two chains run in opposite directions. ..The bases are on the inside of the helix and the phosphates on the outside..."
"The novel feature of the structure is the manner in which the two chains are held together by the purine and pyrimidine bases... The (bases) are joined together in pairs, a single base from one chain being hydrogen-bonded to a single base from the other chain, so that the two lie side by side...One of the pair must be a purine and the other a pyrimidine for bonding to occur. ...Only specific pairs of bases can bond together. These pairs are: adenine (purine) with thymine (pyrimidine), and guanine (purine) with cytosine (pyrimidine)."
"...in other words, if an adenine forms one member of a pair, on either chain, then on these assumptions the other member must be thymine; similarly for guanine and cytosine. The sequence of bases on a single chain does not appear to be restricted in any way. However, if only specific pairs of bases can be formed, it follows that if the sequence of bases on one chain is given, then the sequence on the other chain is automatically determined."
and
"...It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material."
And with these words, the way was made clear for tremendous strides in our understanding of the structure of DNA and, as a result our ability to work with and manipulate the information-rich DNA molecule.
Subscribe to:
Comments (Atom)